Peter Horvath's Portfolio
Publications | Research interest | About | Home

High content
analysis

High content analysis
|
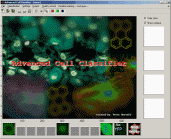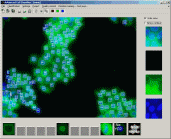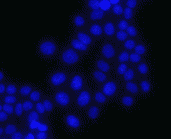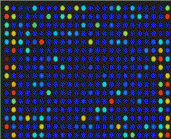
Advanced Cell Classifier - ACC is a data analyser program to evaluate cell-based high-content screens. The basic aim is to provide a very accurate analysis with minimal user interaction using advanced machine learning methods. |
|
|
Genome-scale high content screen analysis - The analysis of the first few Swiss academic whole human genome-wide siRNA screens. The analysis involves quality control, data management, image analysis, feature extraction, cell classification using machine learning, statistical analysis (quality control), and hit detection. |
|
|
Illumination correction of HCS images - My team is interested in developing methods to estimate most accurately vignetting and background functions and correct illumination problems. Our main goal is to estimate the functions via the images and not to use reference objects. Ph.D. project of Filippo Piccinini. |
|
|
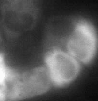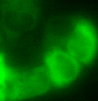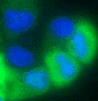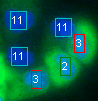
Segmentation and identification of co-cultured cells - We developed a method to detect and distinguish different cell-types in co-cultures. The method is based on a novel spot detection algorithm and classification of the detected spots. |
|
|
Machine learning methods for high content screening (life sciences) - These projects develop methods and software to answer the following questions: Can one suggest the "best" machine learning method for a given biological data set? Can these methods be generally good for other biological data? To what extent the human factor influences the final decisions? How can we introduce novel techniques such as regression, semi-supervised learning, active learning? PosDoc project of Kevin Smith. |
|
|
Statistical methods for screening - We are mainly interested in two fields. Firstly, the normalization and correction of screening data to eliminate biological and liquid handling effects (eg. plate effects, cell number differences, ...). Secondly, to generate reliable and biologically meaningful hit list for replicate experiments or those containing multiple oligo sequences.
|
|
|
|
Microscopic image analysis |
|
Semi-automatic contour tracking - We develope a semi-automated program
using active contours to track ring-like objects (osteoclast cells).
The program also performs temporal, morphological, intensity-, and
texture-based statistics. |
|
|
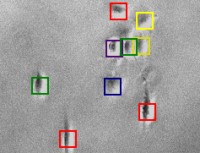
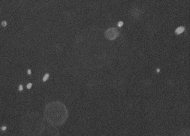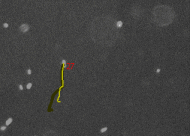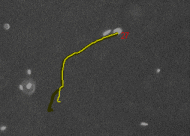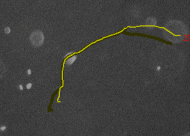
Tracking cells on phase contrast
images - CellTracker is a program to
perform automated and manual cell migration detection. Main goals are:
automated image quality enhancement using background subtraction and
alignment correction; automated detection and tracking of cells;
manual tracking and editing cell paths. Master's thesis of Martin
Maag. |
|
|
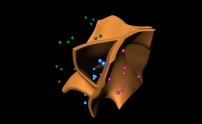
Cell segmentation and splitting in 3D
- We developed methods to split blobs in 3D using gradient vector
flows. Master's thesis of Christoph
Faigle. |
|
|
Statistical analysis of non-sister kinetochore's
motion coupling - We are interested in the
organization of the metaphase plate and the statistical correlation
analysis of non-sister kinetochore migration. Master's thesis of
Katalin Virag. |
|
|
|
|
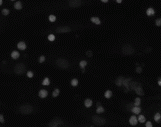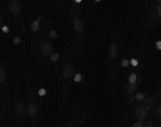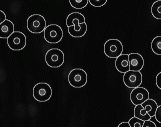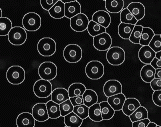
Gas of
Circles Active contour model - Circular object
segmentation. This model is a tool to describe a set of circles with
an approximately fixed radius. The model is based on the
higher-order active contour (HOAC) framework. For certain ranges of
the parameters, the model creates stable circles with an
approximately fixed radius. The images we use are color-infrared
(CIR) and panchromatic aerial images. Experiments show that the
models outperform other traditional circle detection methods. The
model can also be applied to the detection of other circular
objects, e.g. in cell biology, nanotechnology, medical imaging,
satellite images. (INRIA Sophia Antipolis, University of
Szeged). |
|
| 3D tracking on 2D images (BacTrack) - We developed a model to perform 3D tracking on microscopic image sequences using the Point Spread Function of the acquisition system. Master's thesis of Qasim Bukhari. |
|